With a single test, comprehensive genomic profiling (CGP) can analyze a broad panel of genes to detect the four main classes of genomic alterations known to drive cancer growth: base substitutions, insertions and deletions, copy number alterations (CNAs), and rearrangements or fusions. This type of molecular testing produces comprehensive patient reports with a broad and deep assessment of possible underlying oncogenic drivers.
Why Comprehensive Genomic Profiling?
More Insights. Fewer Missed Opportunities.

More and more patients can benefit from molecular profiling due to the rapidly growing number of targeted therapies. These include novel therapies being developed for less prevalent gene alterations like NTRK fusions, which have been identified in less than 1% of all cancers.1 Today, analyzing a broader panel of genes for multiple classes of alterations has become increasingly important.
Understanding the Difference Can Make a Difference.
CGP
Detects the four main classes of genomic alterations across a broader panel of genes
Single Marker
Identifies alterations that are confined to a single gene, potentially missing clinically relevant mutations in additional genes
Hotspot
May test multiple genes at a time, but it is confined to hotspot regions within those genes, resulting in potentially missing other clinically relevant classes of alterations
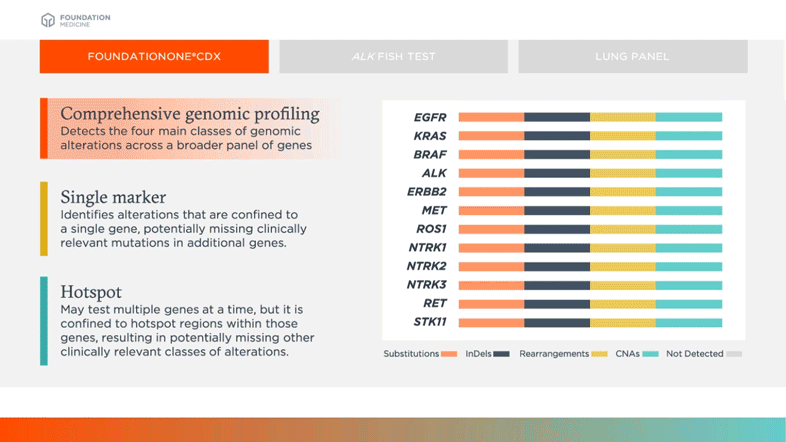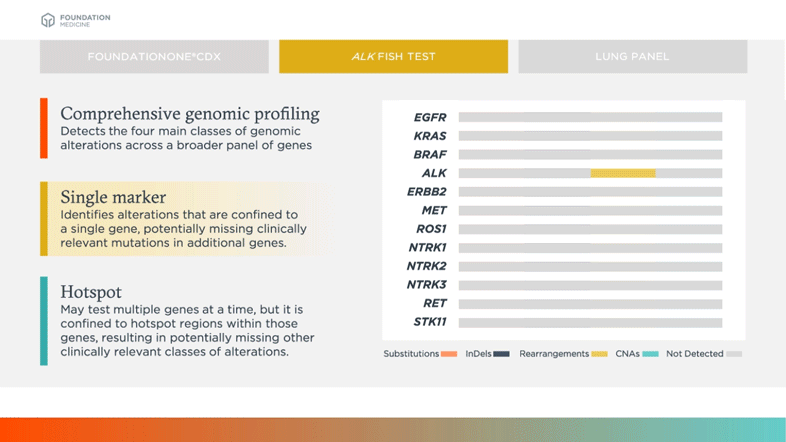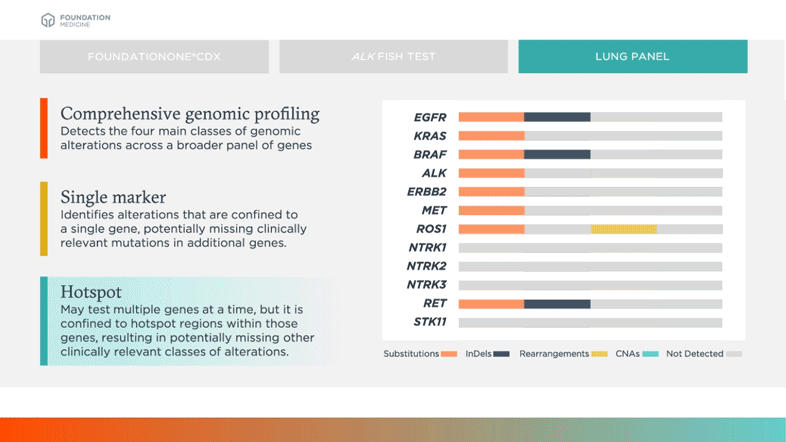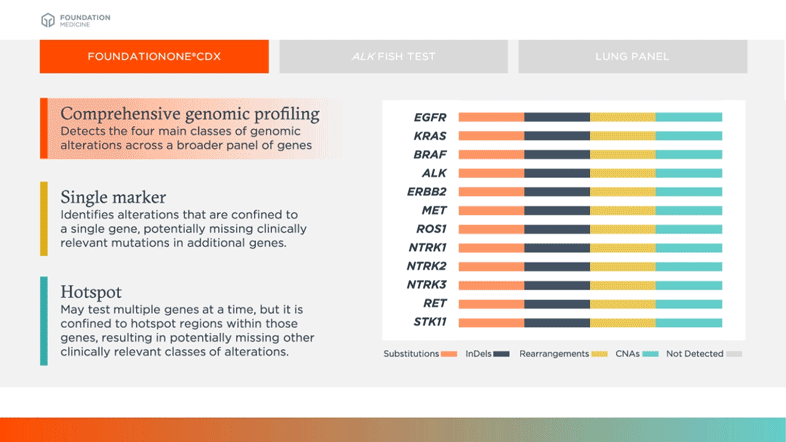
Biomarkers shown in table above are examples of relevant and emerging biomarkers in non-small cell lung cancer (NSCLC). Size is not representative of frequency of alterations.
Common, Complex, and Rare
Comprehensive genomic profiling can provide more complete information on common oncogenic drivers (like EGFR, KRAS, BRAF in NSCLC for example) and new information on complex or rare biomarkers (like MET Exon 14, NTRK1, NTRK2, NTRK3 in NSCLC for example) all from a single test.3,4
Relevant Genomic Signatures
Comprehensive genomic profiling results may include microsatellite instability (MSI) and tumor mutational burden (TMB). Genomic loss of heterozygosity (gLOH) can also be determined through comprehensive genomic profiling.
Efficiency
In some cases, sequential testing of single biomarkers or use of limited molecular diagnostic panels may quickly exhaust sample. Some professional guidelines5,6 recommend that broad molecular profiling be done as part of biomarker testing for eligible patients using a validated test, which can help minimize tissue use and potential wastage.
More Options
For tumor genomic testing in eligible patients, use of tissue is recommended by professional guidelines, with use of plasma ctDNA (liquid biopsy) assays as an option if obtaining sufficient tissue is not clinically feasible.5-7 A study in NSCLC found that 29% of patients didn’t get results from molecular testing because tissue was insufficient.8 In such cases, you need a portfolio that includes liquid-based comprehensive genomic profiling.
Continuously Evolving
Comprehensive genomic profiling can help you keep up to date with the most recent treatment options available throughout the patient’s treatment journey.
Foundation Medicine is Your Trusted Comprehensive Genomic Profiling Partner
Foundation Medicine offers a proven portfolio of comprehensive genomic profiling tests to help identify more treatment options for your advanced cancer patients.
FDA-approved tissue- and blood-based testing is available for all solid tumors with FoundationOne®CDx and FoundationOne®Liquid CDx. In addition, FoundationOne®Heme is a laboratory developed test for hematologic malignancies, sarcomas, or solid tumors where fusion detection is desired.
All Foundation Medicine tests are covered for qualifying Medicare beneficiaries who meet clinical criteria.9
Learn more about our portfolio of CGP tests.
Need More Details?
Our client services team is here to help, Monday through Friday 8AM – 8PM EST.
Additional Notes
NCCN makes no warranties of any kind whatsoever regarding their content, use or application and disclaims any responsibility for their application or use in any way.
1 Fabrizio, et al. Clinical and analytic validation of FoundationOne CDx for NTRK fusion-positive solid tumors in patients treated with entrectinib. In: Proceedings of the AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics; 2019. AACR; Mol Cancer Ther 2019; Abstract A028. doi:10.1158/1535-7163.TARG-19-A028
2 IQVIA Institute for Human Data Science. (August 2020). Supporting Precision Oncology: Targeted Therapies, Immuno-oncology, and Predictive Biomarker-Based Medicines. Retrieved from https://www.iqvia.com/insights/the-iqvia-institute/reports/supporting-precision-oncology
3 Drilon A, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in “driver-negative”lung adenocarcinomas. Clin Cancer Res 2015;21:3631-3639.
4 Rozenblum AB, et al. Clinical impact of hybrid capture-based next-generation sequencing on changes in treatment decisions in lung cancer. J ThoracOncol 2017;12(2):258-68.
5 Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Esophageal and Esophagogastric Junction Cancers V.3.2021.© National Comprehensive Cancer Network, Inc. 2021. All rights reserved. Accessed July 29, 2021. To view the most recent and complete version of the guideline, go online to NCCN.org
6 Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Non-Small Cell Lung Cancer V.5.2021.© National Comprehensive Cancer Network, Inc. 2021. All rights reserved. Accessed July 29, 2021. To view the most recent and complete version of the guideline, go online to NCCN.org.
7 Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Prostate Cancer V.2.2021.© National Comprehensive Cancer Network, Inc. 2021. All rights reserved. Accessed July 29, 2021. To view the most recent and complete version of the guideline, go online to NCCN.org.
8 Kris, M. G. et. al. (2014). Using Multiplexed Assays of Oncogenic Drivers in Lung Cancers to Select Targeted Drugs. JAMA. 2014;311(19):1998-2006.
9 For FoundationOne®CDx and FoundationOne®Liquid CDx, see “Decision for Next Generation Sequencing (NGS) for Medicare Beneficiaries with Advanced cancer – CAG-00450R.” (See Appendix B). For FoundationOne®Heme, see the “Local Coverage Determination (LCD): MolDX: NEXT-GENERATION Sequencing Lab-Developed Tests for Myeloid Malignancies and Suspected Myeloid Malignancies (L38047)”.
Important Safety Information
FoundationOne®CDx and FoundationOne®Liquid CDx are qualitative next-generation sequencing based in vitro diagnostic tests for advanced cancer patients with solid tumors and are for prescription use only. FoundationOne CDx utilizes FFPE tissue and analyzes 324 genes as well as genomic signatures. FoundationOne Liquid CDx analyzes 324 genes utilizing circulating cell-free DNA and is FDA-approved to report short variants in 311 genes. The tests are companion diagnostics to identify patients who may benefit from treatment with specific therapies in accordance with the therapeutic product labeling. Additional genomic findings may be reported and are not prescriptive or conclusive for labeled use of any specific therapeutic product. Use of the tests does not guarantee a patient will be matched to a treatment. A negative result does not rule out the presence of an alteration. Some patients may require a biopsy for testing with FoundationOne CDx when archival tissue is not available which may pose a risk. When considering eligibility for ROZLYTREK® based on the detection of NTRK1/2/3 and ROS1 fusions, or for TEPMETKO® based on the detection of MET SNVs and indels that lead to MET exon 14 skipping, testing using plasma specimens is only appropriate for patients for whom tumor tissue is not available for testing. Patients who are tested with FoundationOne Liquid CDx and are negative for other companion diagnostic mutations should be reflexed to tumor tissue testing and mutation status confirmed using an FDA-approved tumor tissue test, if feasible. For the complete label, including companion diagnostic indications and important risk information, please visit www.F1CDxLabel.com and www.F1LCDxLabel.com.